Morphologically and Anatomically accuRate Segmentation (MARS)
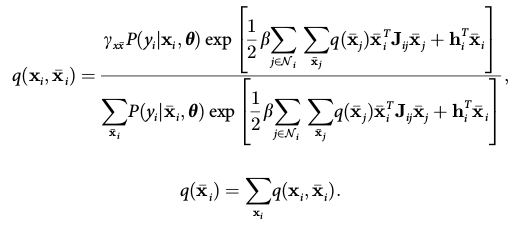
Currently available automated segmentation tools only provide results for brain tissues (gray matter, white matter, cerebrospinal fluid (CSF)), have a limited FOV, and do not guarantee continuity and smoothness of tissues, which is crucially important for accurate modeling of transcranial electrical or magnetic stimulation. MARS is a tool that addresses these needs.
MARS is an extended toolbox for SPM software. It is developed based on the SPM8 toolbox "New Segment", which is an implementation of the Unified Segmentation algorithm (Ashburner & Friston 2005), where the image intensity model and anatomical prior (atlas) are combined into a Bayesian inference framework. MARS further added the morphological constraints on neighboring tissue voxels to the framework, which is encoded by Markov Random Field (MRF). This MRF information is added into the updating equation of the E-step of the algorithm, not just as a post-processing step. Compared to the New Segment, the resulting segmentation is shown to be more smooth and with less discontinuities, while at the same time the accuracy is not significantly changed.
MARS is always initialized by the New Segment. In other words, this toolbox will run New Segment in the first step to get the optimal estimates on the bias field, the registration parameters between atlas and the image(s) to be segmented, and the initial estimates of segmentation posteriors and parameters of Gaussian mixture model. MARS will then continues to update the segmentation posteriors and Gaussian mixture model by using both atlas (tissue probability map, TPM) and tissue correlation map (TCM), while keep the bias field and registration parameters untouched. The toolbox is backwards compatible with the New Segment toolbox, as users can disable the MARS update so that only New Segment will be run.
The TCM is a way of representing MRF constraint. It indicates the probability of co-occurrence of specific two tissues in two neighboring voxels. For example, white matter cannot be adjacent to the air, and thus the probability of observing white matter as a neighbor of air voxel is 0, and this is encoded in the TCM. We developed three categories of TCM: the local, global and regional. The local TCM gives different co-occurrence probabilities depending on the location of the voxels, while the global version uses the same value for all the voxels in the image. The local TCM has nice theoretical properties, but not practical for implementation, as it needs too much memory to store and compute; while the global one is too rough to include any local information. The regional TCM is a trade-off between the two: it encodes locality information such as the interior of white matter and the boundary between gray and white matters, while at the same time uses the same co-occurrence value for one specific locality. Therefore, the regional TCM is highly recommended, and local TCM is not recommended unless you have a 64-bit computer with at least 50 GB memory.
For more details on the algorithm, see Yu Huang, Lucas C. Parra, Fully Automated Whole-Head Segmentation with Improved Smoothness and Continuity, with Theory Reviewed. PLoS ONE 10(5): e0125477.
Download the Matlab code and TPM/TCM here. Note MARS is only compatible with SPM8, not with SPM12.
Download and use of this work is subject to this license.